Presented by Emi Tanaka
School of Mathematics and Statistics
dr.emi.tanaka@gmail.com
@statsgen
1st Dec 2019 @ Biometrics by the Botanic Gardens | Adelaide, Australia
🔎 Open and inspect the file
demo-header.Rmd
Cross Reference
- When you make a header via Rmd
- The id is created by replacing space with
-and making it all lower case. - Now you can link to this header by
[some text](#some-header). - Cross references work for both pdf and html outputs.
Direct Reference for html
- For
htmloutput, you can also give a link directly to the relevant section - E.g. open
demo-header.htmlin a web browser - Append say
#chicken-datato the url. It should look likedemo-header.html#chicken-data - It should have taken you to straight to the corresponding header 🏃
User-defined id
- You can define your own id by appending
{#your-id}.
# Some header {#header1}- Now you can link to this header with the id
header1. - Note there should be no space in the id name!
🔍 Open and inspect the file
demo.bib
Bibliography
- BibTeX citation style format is used to store references in
.bibfiles. - Remember that you can get most BibTeX citation for R packages
citationfunction. (Scroll below to see the BibTeX citation).
citation("xaringan") To cite package 'xaringan' in publications use: Yihui Xie (2019). xaringan: Presentation Ninja. R package version 0.9. https://CRAN.R-project.org/package=xaringan A BibTeX entry for LaTeX users is @Manual{, title = {xaringan: Presentation Ninja}, author = {Yihui Xie}, year = {2019}, note = {R package version 0.9}, url = {https://CRAN.R-project.org/package=xaringan}, }🔍 Open, inspect and knit the file
demo-citation.Rmd
Citations
- You can include BibTeX by specifying the
bibfile at YAML as:
bibliography: bibliography.bib[@bibtex-key] (Author et al. 2019)
or
@bibtex-key Author et al. 2019
- See
demo-citation.Rmd
Figure References
- Support for figure references are included for output format type
bookdown::pdf_document2for pdf orbookdown::html_document2for html.
```{r plot1, fig.cap = "Caption"}ggplot(cars, aes(dist, speed)) + geom_point()```Above figure number can be referenced as
\@ref(fig:plot1)The reference label has the prefix
fig:before the chunk label.
Table References
- Support for table references are also included for output format type
bookdown::pdf_document2for pdf orbookdown::html_document2for html.
```{r table1}knitr::kable(cars, booktabs = TRUE, caption = "Caption")```Above table number can be referenced as
\@ref(tab:table1)The reference label has the prefix
tab:before the chunk label.
Markdown for Captions
```{r plot1, fig.cap = "(ref:label)"}ggplot(cars, aes(dist, speed)) + geom_point()```- Then the caption can be entered in a separate paragraph with empty lines above and below it
(ref:label) This is the *caption* with **markdown**.- You can substitute
labelwith another unique label composed of alphanumeric characters, :, -, or / - This caption supports markdown syntax
- This is great for long captions
- It also works for tables!
🔧 Open and work through
challenge-12-references.Rmd
05:00
Parametrized Report
---title: "Parameterized Report"params: species: setosa output: html_document---```{r, message = FALSE, fig.dim = c(3,2)}library(tidyverse)iris %>% filter(Species==params$species) %>% ggplot(aes(Sepal.Length, Sepal.Width)) + geom_point(aes(color=Species))```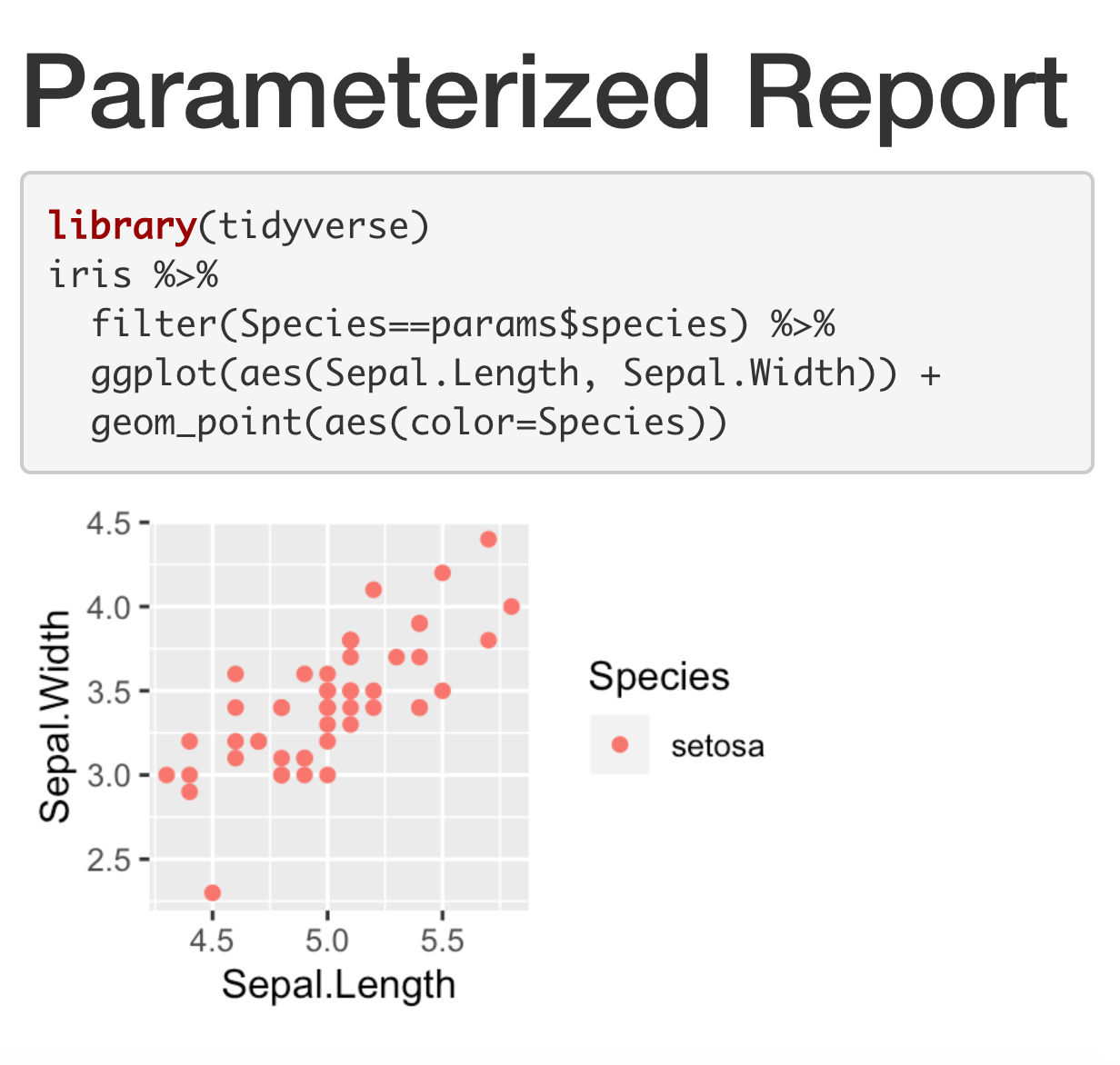
Knit with Parameters
---title: "Parameterized Report"params: species: label: "Species" value: setosa input: select choices: [setosa, versicolor, virginica] color: red max: label: "Maximum Sepal Width" value: 4 input: slider min: 4 max: 5 step: 0.1output: html_document---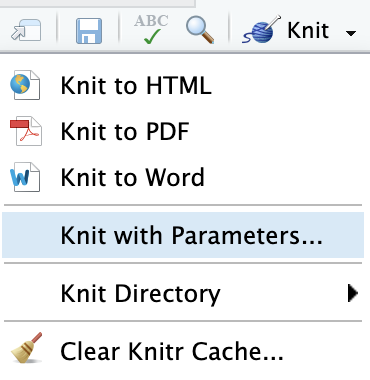
```{r, message = params$printmsg}library(tidyverse)iris %>% filter(Species==params$species) %>% filter(Sepal.Width < params$max) %>% ggplot(aes(Sepal.Length, Sepal.Width)) + geom_point(color = params$color) + labs(title = params$species)```Shiny Report Generator
---title: "Parameterized Report"params: species: label: "Species" value: setosa input: select choices: [setosa, versicolor, virginica] color: red max: label: "Maximum Sepal Width" value: 5 input: slider min: 4 max: 5 step: 0.05output: html_document---
🔧 Open and work through
challenge-13-params.Rmd
05:00
R Markdown via Command Line
demo-render.Rmd
---title: "Parameterized Report"params: species: setosaoutput: html_document---```{r, message = FALSE, fig.dim = c(3,2)}library(tidyverse)iris %>% filter(Species==params$species) %>% ggplot(aes(Sepal.Length, Sepal.Width)) + geom_point(aes(color=Species))```You can knit this file via R command by
using the render function:
library(rmarkdown)render("demo-render.Rmd")You can overwrite the YAML values
by supplying arguments to render:
library(rmarkdown)render("demo-render.Rmd", output_format = "pdf_document", params = list(species = "virginica"))🔧 Open and work through
challenge-14-letters.Rmd
10:00
Themes: html_document
You can change the look of the html document by specifying themes:
default
cerulean
journal
flatly
darkly
readable
spacelab
united
cosmo
lumen
paper
sandstone
simplex
yeti
NULL
output: html_document: theme: ceruleanThese bootswatch themes attach the whole bootstrap library which makes your html file size larger.
prettydoc
prettydoc 📦 is a community contributed theme that is light-weight:
cayman
tactile
architect
leonids
hpstr
rmdformats
rmdformats 📦 contains four built-in html formats:
readthedown
html_clean
html_docco
material
You can use these formats by simply specifying the output in YAML as below:
output: rmdformats::readthedownSee more about it below:
rticles - LaTeX Journal Article Templates
acm
acs
aea
agu
amq
ams
asa
biometrics
copernicus
elsevier
frontiers
ieee
jss
mdpi
mnras
peerj
plos
pnas
rjournal
rsos
rss
sage
sim
springer
tf
External Files in Templating
- When using
rticles, each journal usually require external files (e.g.clsor image files). - These external components are stored within the package.
- So use
draftinstead ofrender!
GUI
RStudio > File > New File > R Markdown ... > From Template
Command line
rmarkdown::draft("file.Rmd", template = "biometrics_article", package = "rticles")Making your own R Markdown template
- You need to make an R package first!
Go to RStudio > New Project > New Directory > R Package orusethis::create_package()
- When you are in your R package project,
usethis::use_rmarkdown_template("<Name>")- Modify the
skeleton/skeleton.Rmdto how you want and add all external files to theskeletonfolder. - Install your package.
- 🎉 And now find it at RStudio > File > New File > R Markdown > From Template.
🔧 Create your own
R Markdown Template Package!
Session Information
devtools::session_info() ─ Session info ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── setting value version R version 3.6.0 (2019-04-26) os macOS Mojave 10.14.6 system x86_64, darwin15.6.0 ui X11 language (EN) collate en_AU.UTF-8 ctype en_AU.UTF-8 tz Australia/Adelaide date 2019-12-03 ─ Packages ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── package * version date lib source anicon 0.1.0 2019-05-28 [1] Github (emitanaka/anicon@377aece) assertthat 0.2.1 2019-03-21 [1] CRAN (R 3.6.0) backports 1.1.4 2019-04-10 [1] CRAN (R 3.6.0) broom 0.5.2 2019-04-07 [1] CRAN (R 3.6.0) callr 3.3.1 2019-07-18 [1] CRAN (R 3.6.0) cellranger 1.1.0 2016-07-27 [1] CRAN (R 3.6.0) cli 1.1.0 2019-03-19 [1] CRAN (R 3.6.0) colorspace 1.4-1 2019-03-18 [1] CRAN (R 3.6.0) countdown 0.2.0 2019-05-30 [1] Github (gadenbuie/countdown@c8e8710) crayon 1.3.4 2017-09-16 [1] CRAN (R 3.6.0) desc 1.2.0 2018-05-01 [1] CRAN (R 3.6.0) devtools 2.0.2 2019-04-08 [1] CRAN (R 3.6.0) digest 0.6.22 2019-10-21 [1] CRAN (R 3.6.0) dplyr * 0.8.3 2019-07-04 [1] CRAN (R 3.6.0) emo 0.0.0.9000 2019-06-03 [1] Github (hadley/emo@02a5206) evaluate 0.14 2019-05-28 [1] CRAN (R 3.6.0) forcats * 0.4.0 2019-02-17 [1] CRAN (R 3.6.0) fs 1.3.1 2019-05-06 [1] CRAN (R 3.6.0) generics 0.0.2 2018-11-29 [1] CRAN (R 3.6.0) ggplot2 * 3.2.1 2019-08-10 [1] CRAN (R 3.6.0) glue 1.3.1.9000 2019-10-24 [1] Github (tidyverse/glue@71eeddf) gtable 0.3.0 2019-03-25 [1] CRAN (R 3.6.0) haven 2.1.0 2019-02-19 [1] CRAN (R 3.6.0) hms 0.5.1 2019-08-23 [1] CRAN (R 3.6.0) htmltools 0.4.0 2019-10-04 [1] CRAN (R 3.6.0) httr 1.4.1 2019-08-05 [1] CRAN (R 3.6.0) icon 0.1.0 2019-05-28 [1] Github (ropenscilabs/icon@a510f88) jsonlite 1.6 2018-12-07 [1] CRAN (R 3.6.0) knitr 1.25 2019-09-18 [1] CRAN (R 3.6.0) lattice 0.20-38 2018-11-04 [1] CRAN (R 3.6.0) lazyeval 0.2.2 2019-03-15 [1] CRAN (R 3.6.0) lifecycle 0.1.0 2019-08-01 [1] CRAN (R 3.6.0) lubridate 1.7.4 2018-04-11 [1] CRAN (R 3.6.0) magrittr 1.5 2014-11-22 [1] CRAN (R 3.6.0) memoise 1.1.0 2017-04-21 [1] CRAN (R 3.6.0) modelr 0.1.4 2019-02-18 [1] CRAN (R 3.6.0) munsell 0.5.0 2018-06-12 [1] CRAN (R 3.6.0) nlme 3.1-140 2019-05-12 [1] CRAN (R 3.6.0) pillar 1.4.2 2019-06-29 [1] CRAN (R 3.6.0) pkgbuild 1.0.3 2019-03-20 [1] CRAN (R 3.6.0) pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 3.6.0) pkgload 1.0.2 2018-10-29 [1] CRAN (R 3.6.0) prettyunits 1.0.2 2015-07-13 [1] CRAN (R 3.6.0) processx 3.4.1 2019-07-18 [1] CRAN (R 3.6.0) ps 1.3.0 2018-12-21 [1] CRAN (R 3.6.0) purrr * 0.3.2 2019-03-15 [1] CRAN (R 3.6.0) R6 2.4.0 2019-02-14 [1] CRAN (R 3.6.0) Rcpp 1.0.2 2019-07-25 [1] CRAN (R 3.6.0) readr * 1.3.1 2018-12-21 [1] CRAN (R 3.6.0) readxl 1.3.1 2019-03-13 [1] CRAN (R 3.6.0) remotes 2.0.4 2019-04-10 [1] CRAN (R 3.6.0) rlang 0.4.0.9000 2019-08-03 [1] Github (r-lib/rlang@b0905db) rmarkdown 1.16 2019-10-01 [1] CRAN (R 3.6.0) rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.6.0) rstudioapi 0.10 2019-03-19 [1] CRAN (R 3.6.0) rvest 0.3.4 2019-05-15 [1] CRAN (R 3.6.0) scales 1.0.0 2018-08-09 [1] CRAN (R 3.6.0) sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 3.6.0) stringi 1.4.3 2019-03-12 [1] CRAN (R 3.6.0) stringr * 1.4.0 2019-02-10 [1] CRAN (R 3.6.0) testthat 2.2.1 2019-07-25 [1] CRAN (R 3.6.0) tibble * 2.1.3 2019-06-06 [1] CRAN (R 3.6.0) tidyr * 1.0.0 2019-09-11 [1] CRAN (R 3.6.0) tidyselect 0.2.5 2018-10-11 [1] CRAN (R 3.6.0) tidyverse * 1.2.1 2017-11-14 [1] CRAN (R 3.6.0) usethis 1.5.0 2019-04-07 [1] CRAN (R 3.6.0) vctrs 0.2.0.9000 2019-08-03 [1] Github (r-lib/vctrs@11c34ae) whisker 0.3-2 2013-04-28 [1] CRAN (R 3.6.0) withr 2.1.2 2018-03-15 [1] CRAN (R 3.6.0) xaringan 0.9 2019-03-06 [1] CRAN (R 3.6.0) xfun 0.10 2019-10-01 [1] CRAN (R 3.6.0) xml2 1.2.0 2018-01-24 [1] CRAN (R 3.6.0) yaml 2.2.0 2018-07-25 [1] CRAN (R 3.6.0) zeallot 0.1.0 2018-01-28 [1] CRAN (R 3.6.0) [1] /Library/Frameworks/R.framework/Versions/3.6/Resources/libraryThese slides are licensed under