# Header 1## Header 2### Header 3#### Header 4##### Header 5###### Header 6R Markdown Basics with
Presented by Emi Tanaka
School of Mathematics and Statistics
dr.emi.tanaka@gmail.com
@statsgen
1st Dec 2019 @ Biometrics by the Botanic Gardens | Adelaide, Australia
What you can do with R Markdown?
What you can do with R Markdown?
What you can do with R Markdown?
What you can do with R Markdown?
What you can do with R Markdown?
Available at https://otexts.com/fpp2/
The pdf book version is also made using R Markdown.
What you can do with R Markdown?
What you can do with R Markdown?
Possibilities are endless...
- Microsoft Word document (.doc, .docx)
- Open Document Text (.odt)
- Rich text format (.rtf)
- Power point slides (.ppt, .pptx)
- Posters (as .html or .pdf)
- Resume (as .html or .pdf)
- Interactive R Notebooks (.html)
- Markdown documents (.md)
- Dashboard (.html)
Before R Markdown (and Sweave)
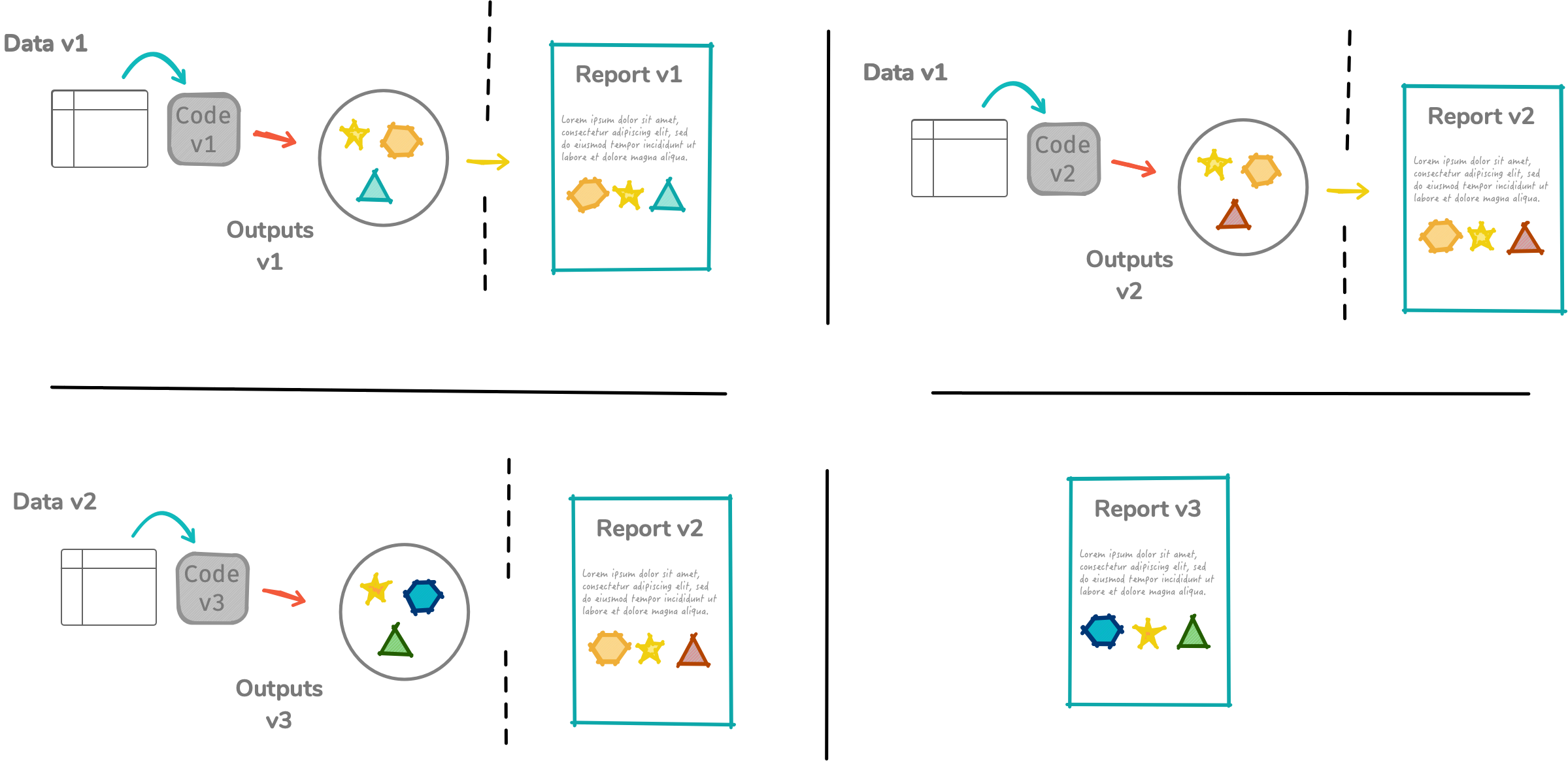
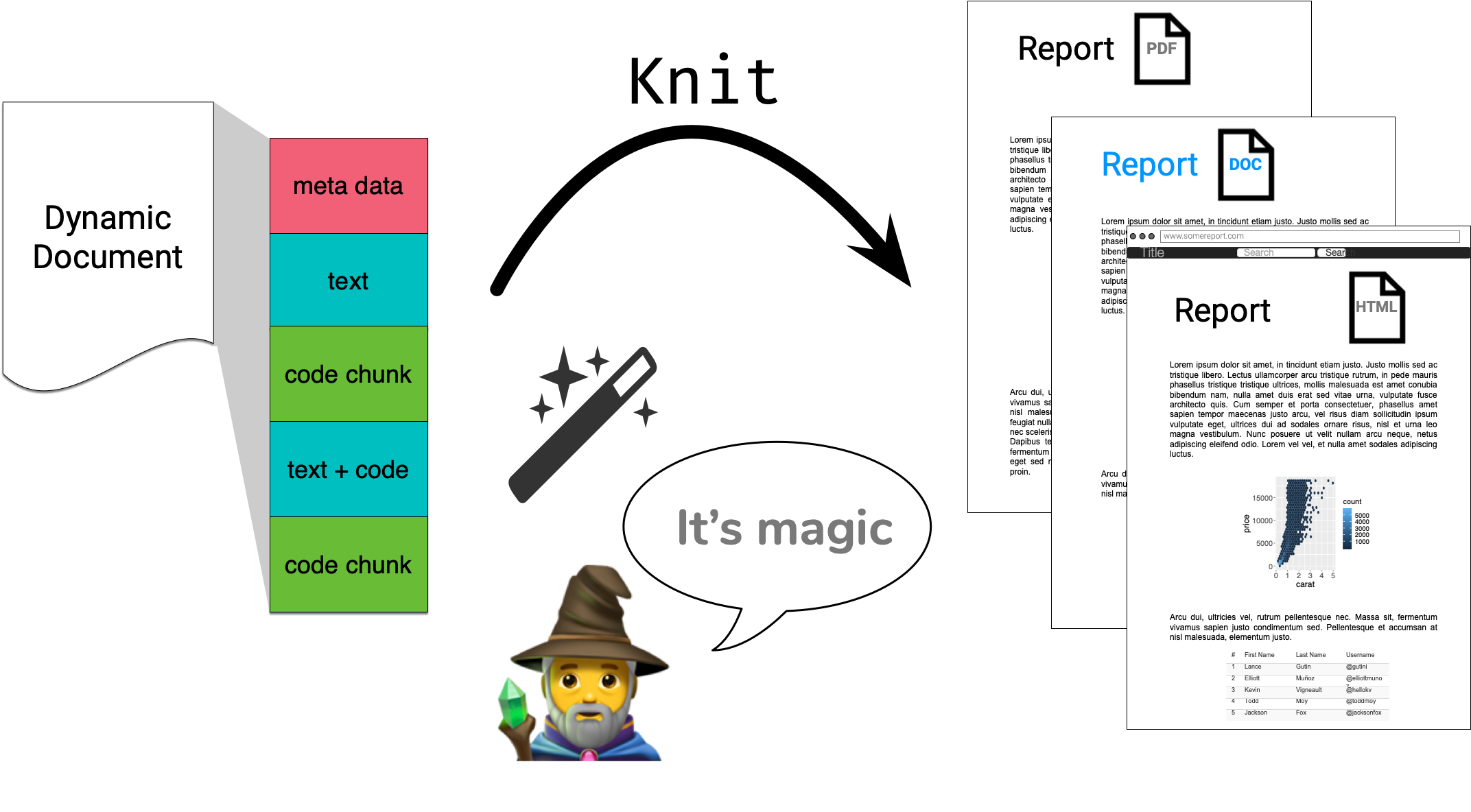
R Markdown in a nutshell 🥜
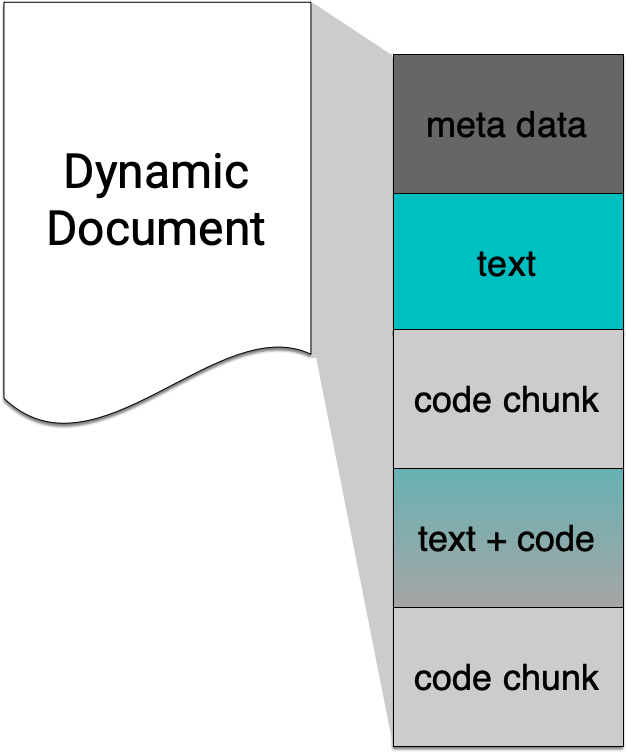
R Markdown integrates text + code in one source document with ability to knit to many output formats (via Pandoc).
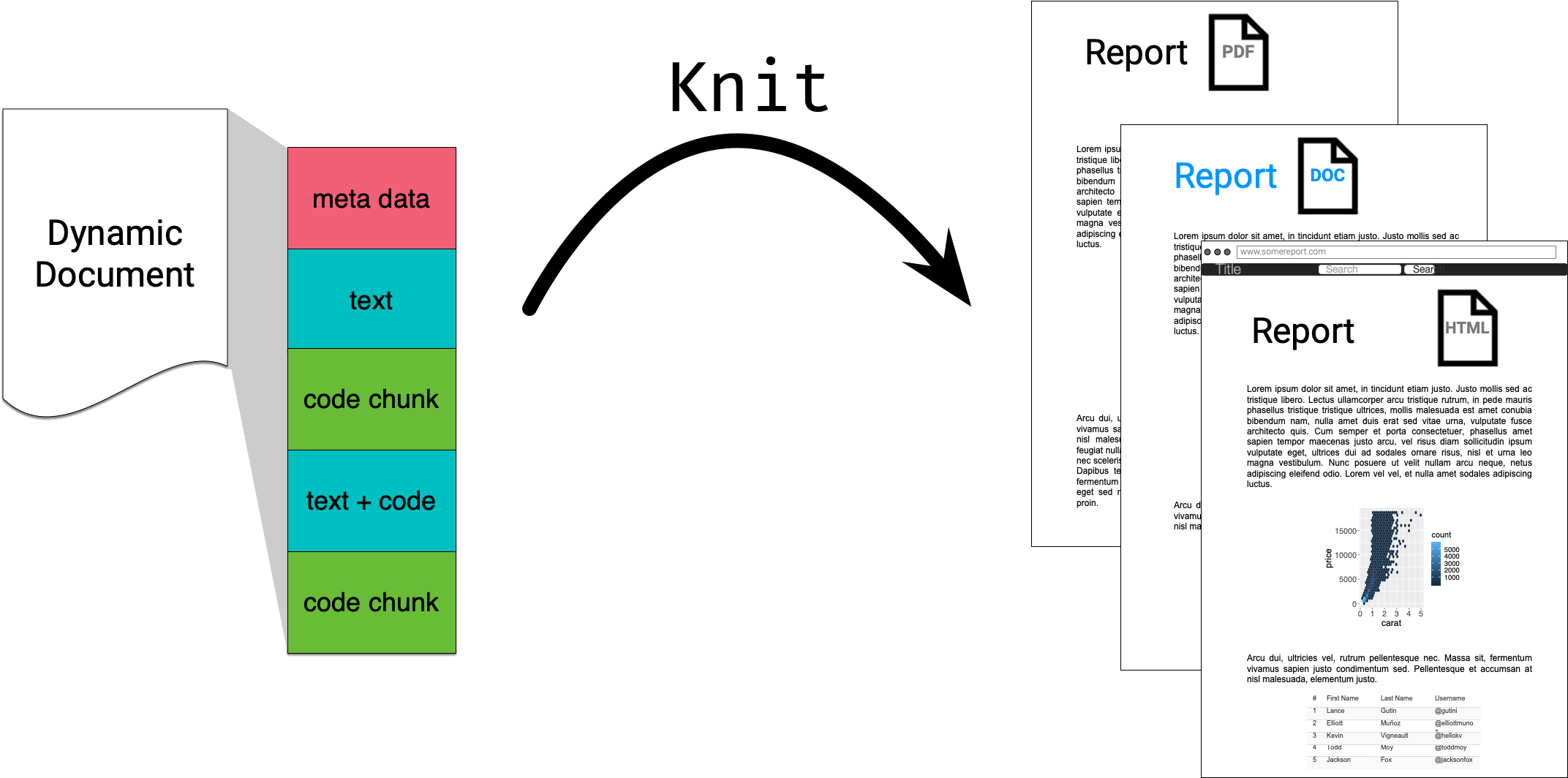
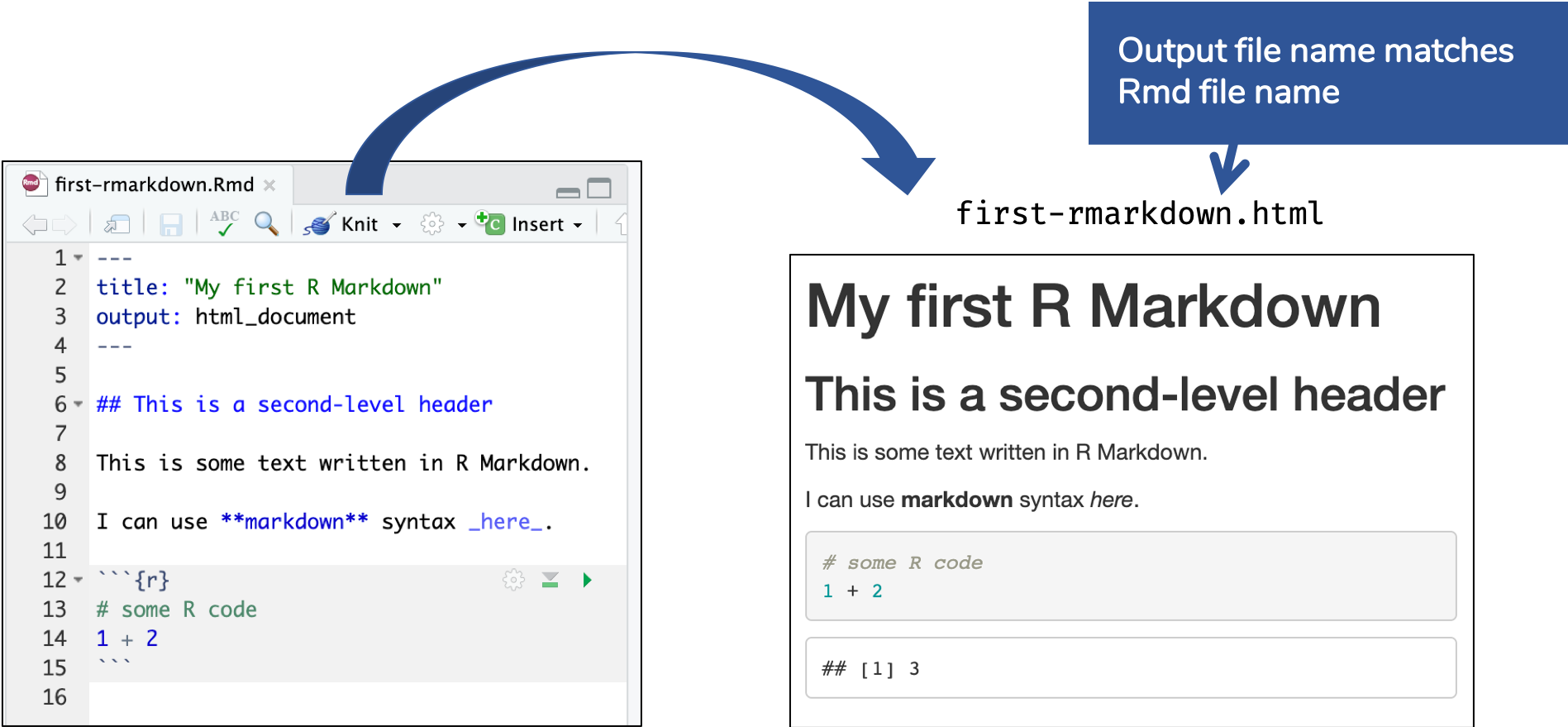
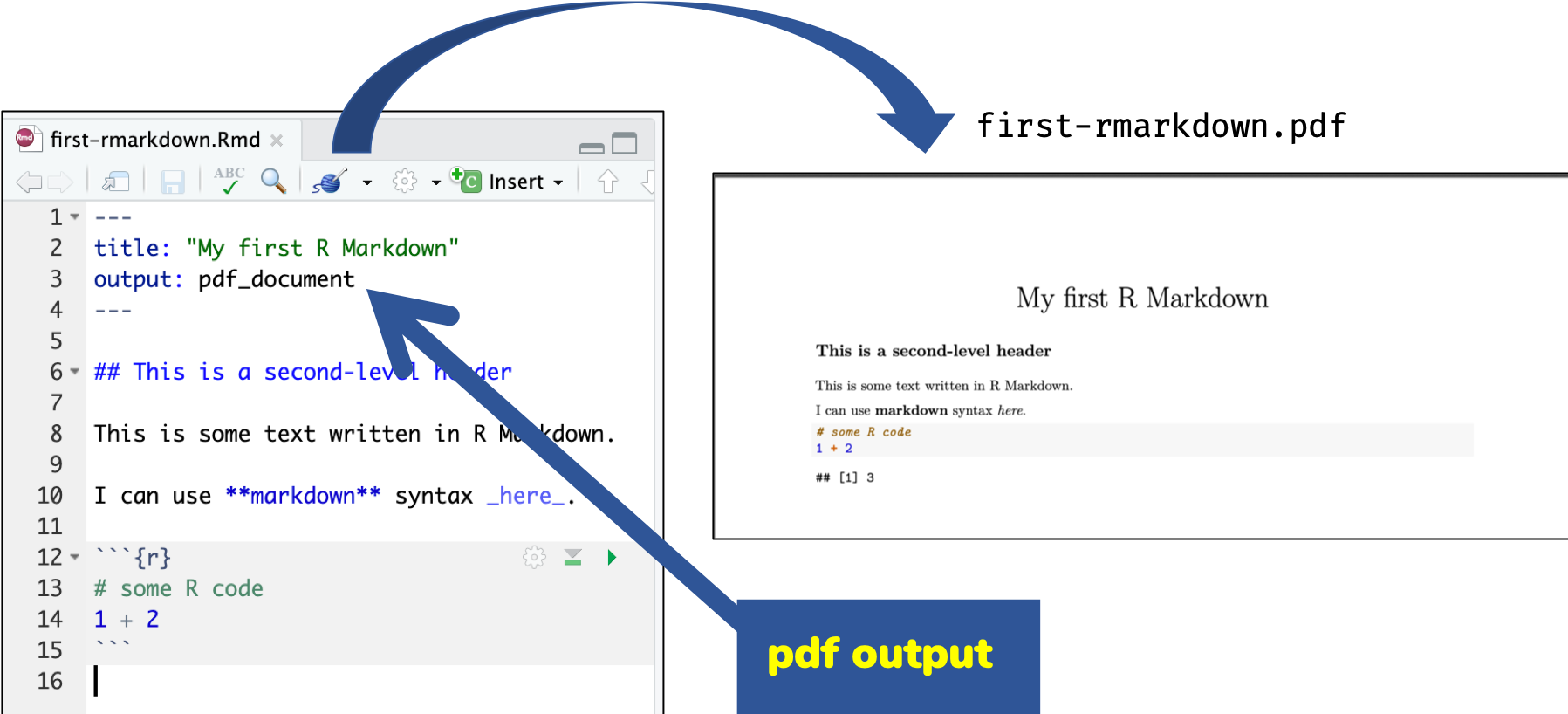
🔍 Open and inspect the file
first-rmarkdown.Rmd
R Markdown Basics
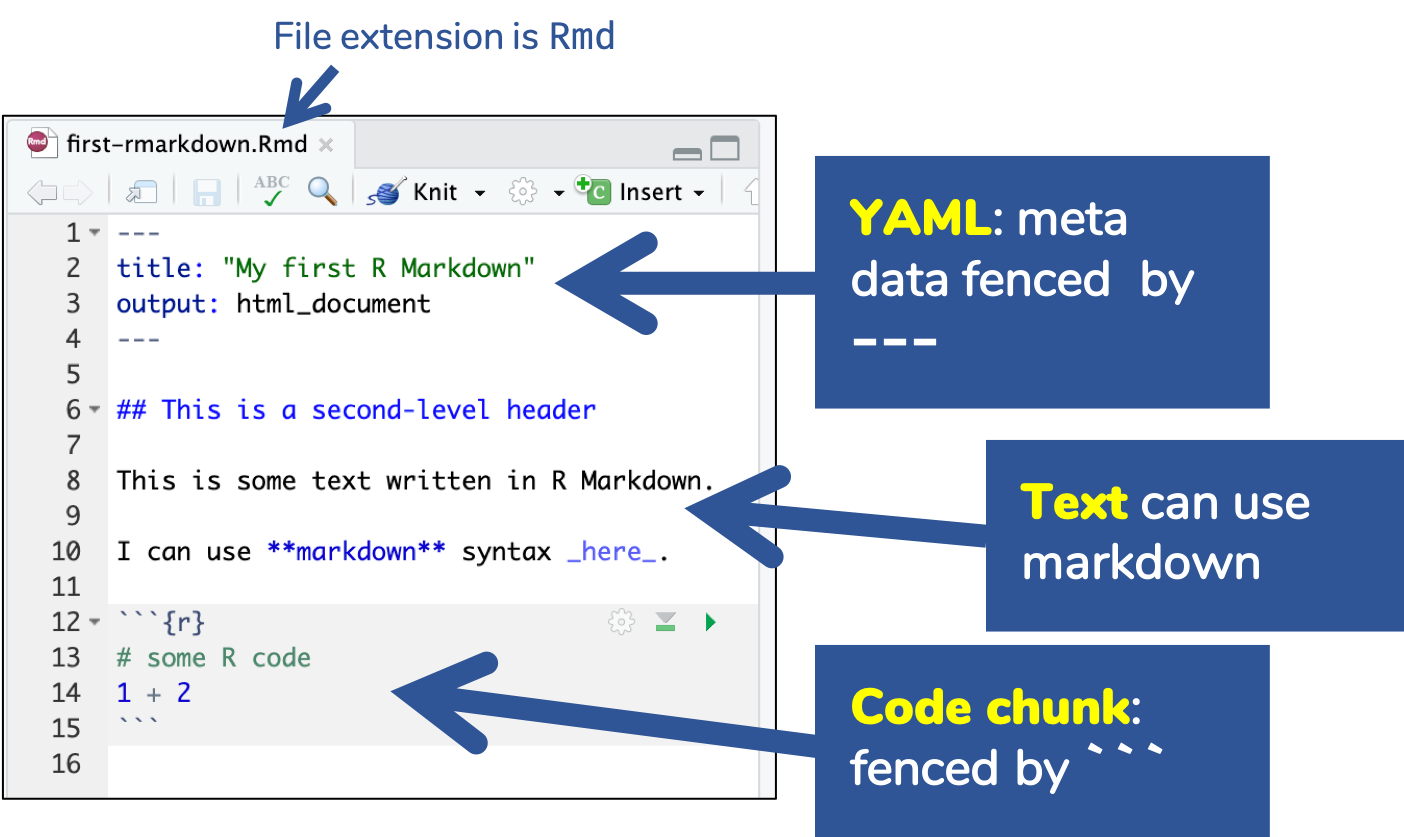
Knitting: Rmd md html
Knitting: Rmd md pdf
How does it all work?
R Markdown under the hood: html_document
knitr 📦 does the hard work of converting Rmd md
---title: "My Report"output: html_document---# Hello```{r}1 + 1```---title: "My Report"output: html_document---# Hello```r1 + 1``````## [1] 2```Rmd
md
R Markdown under the hood: html_document
Then rmarkdown 📦 prepares md for pandoc to convert to html .
---title: "My Report"output: html_document---# Hello```r1 + 1``````## [1] 2```<body><div class="container-fluid main-container"><div class="fluid-row" id="header"><h1 class="title toc-ignore">My Report</h1></div><div id="hello" class="section level1"><h1>Hello</h1><pre class="r"><code>1 + 1</code></pre><pre><code>## [1] 2</code></pre></div></div></body>md
html
R Markdown under the hood: pdf_document
Same as before: knitr 📦 converts Rmd md
---title: "My Report"output: pdf_document---# Hello```{r}1 + 1```---title: "My Report"output: pdf_document---# Hello```r1 + 1``````## [1] 2```Rmd
md
R Markdown under the hood: pdf_document
Then rmarkdown 📦 prepares md for pandoc to convert to tex pdf .
---title: "My Report"output: html_document---# Hello```r1 + 1``````## [1] 2```\begin{document}\maketitle\hypertarget{hello}{%\section{Hello}\label{hello}}\begin{Shaded}\begin{Highlighting}[]\DecValTok{1} \OperatorTok{+}\StringTok{ }\DecValTok{1}\end{Highlighting}\end{Shaded}\begin{verbatim}## [1] 2\end{verbatim}\end{document}md
tex
\begin{document} due to restriction of space.
Meta data with YAML
YAML - YAML Ain't Markup Language
Basic format
---key: value---Example
---title: "R Markdown Workshop"subtitle: "The Basics"author: "Emi Tanaka"date: "`r Sys.Date()`"output: html_document---There must be a space after ":"!
Default (minimal) html output
<!DOCTYPE html><html xmlns="http://www.w3.org/1999/xhtml"><head><meta name="author" content="Emi Tanaka" /><meta name="date" content="2019-11-19" /><title>R Markdown Workshop</title></head><body><h1 class="title toc-ignore">R Markdown Workshop</h1><h3 class="subtitle">The Basics</h3><h4 class="author">Emi Tanaka</h4><h4 class="date">2019-11-19</h4></body></html>html meta data
Default html template add special YAML key values to file automatically

YAML structure
- White spaces indicate structure in YAML - don't use tabs though!
- Same as R, you can comment lines by starting with
#. - YAML is case sensitive.
- A
keycan hold multiple values.
key: - value1 - value2key: [value1, value2]YAML with multiple key values
---title: "R Package & R Markdown Workshop"author: - "Damjan Vukcevic" - "Emi Tanaka"output: html_document---html raw file
<body><h1 class="title toc-ignore">R Package & R Markdown Workshop</h1><h4 class="author">Damjan Vukcevic</h4><h4 class="author">Emi Tanaka</h4> </body>
Values spanning multiple lines
---title: > this is a **single line**abstract: | this value spans *many lines* and appears as it is output: pdf_document---Note that there are two spaces after each line in the above YAML values. This is necessary to create a new line for markdown.
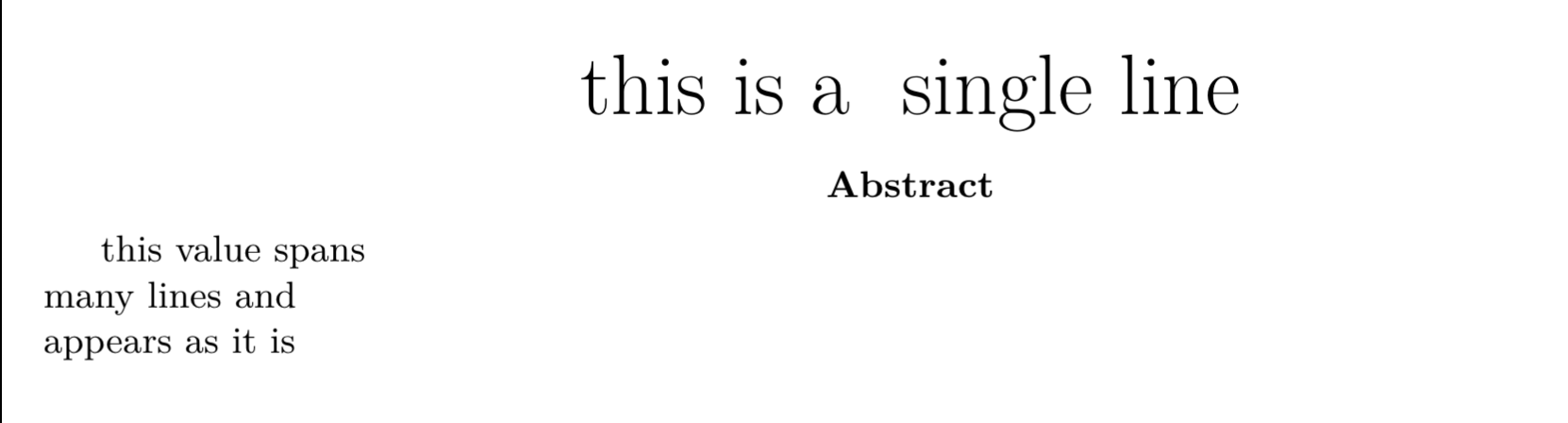
🔍 Open and inspect the file
second-rmarkdown.Rmd
key can contain keys
---output: html_document: toc: true toc_float: true code_download: true code_folding: "hide"---What does each of the above keys do?
04:00
ymlthis 📦
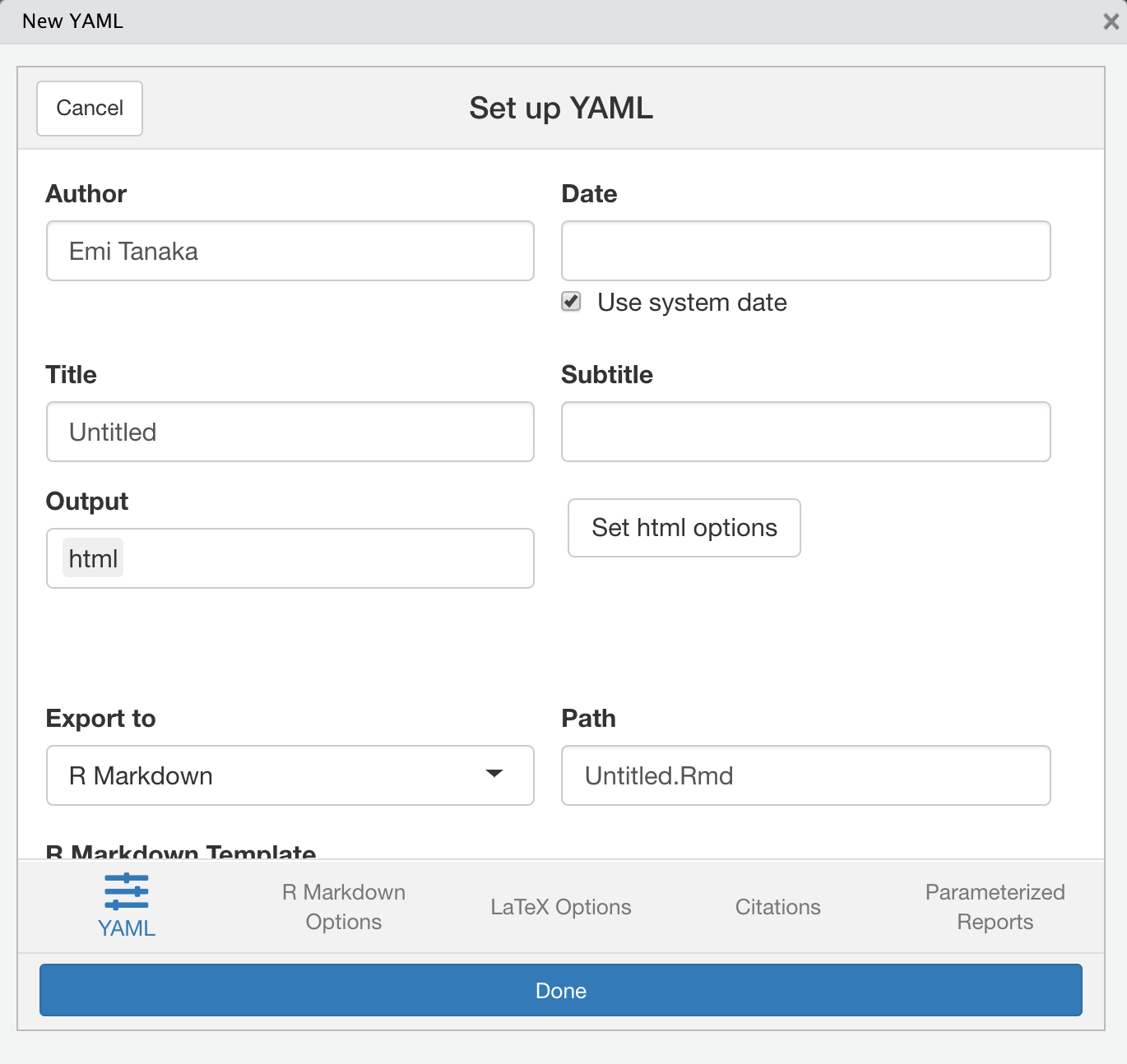
- If you installed
ymlthisthen go to RStudio > Addins > YMLTHIS - It will launch a shiny app with various YAML options
- See also helper functions at https://ymlthis.r-lib.org/
🔧 Open and inspect the file
challenge-05-yaml.Rmd
Text
Text in Markdown: Headers
Header 1
Header 2
Header 3
Header 4
Header 5
Header 6
Text in Markdown: Formatting
**This text is bold** __This text is also bold__ *This text is italic* _This text is also italic_ **_You can combine both_**This text is bold
This text is also bold
This text is italic
This text is also italic
You can combine both
Text in Markdown: Lists
1. Breakfast * Cereal * Milk 1. Lunch * Pizza * Tissue box * Soft * Hard * Soy sauce- Breakfast
- Cereal
- Milk
- Lunch
- Pizza
- Tissue box
- Soft
- Hard
- Soy sauce
Markdown: lightweight markup language
Focus on writing - add flair later
markdown
# Section Bla bla.Then **bold**.html
<h1>Section</h1>Bla bla.Then <b>bold</b>.latex
\section{Section}Bla bla.Then \textbf{bold}.docx
Section
Bla bla.
Then bold.
🔧 Open and go through:
challenge-06-table.Rmd
05:00
Code chunk
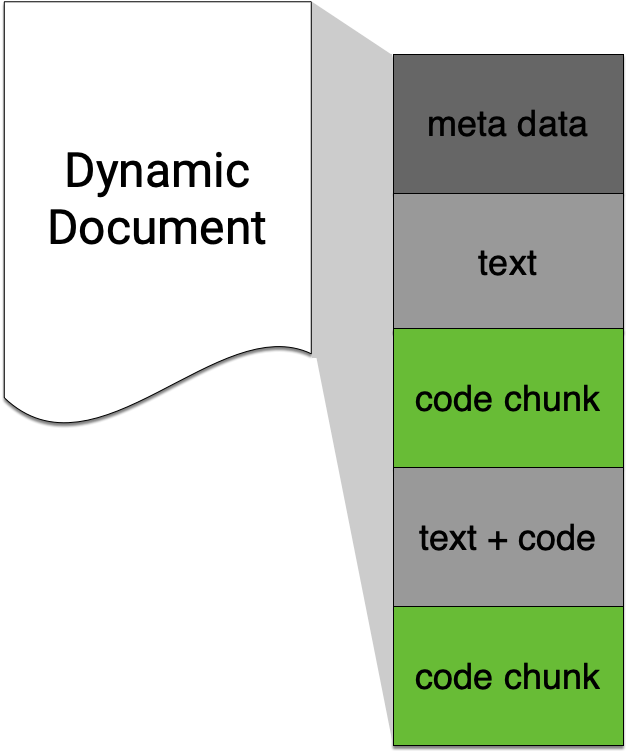
Keyboard shortcut for inserting code chunk
In RStudio .Rmd press
- Mac: ⌘ + ⌥ + i
- PC: Ctrl + Alt + i
to insert a chunk of R code
```{r}```Chunk options: echo & eval
```{r, echo = FALSE}plot(speed ~ dist, cars)``````{r, eval = FALSE}plot(speed ~ dist, cars)```plot(speed ~ dist, cars)Valid chunk options
- Chunk options must be written in one line, i.e. no line break.
- All option values must be valid R expressions. Exception is the chunk name. E.g.
fig.path = figures/is not valid butfig.path = "figures/"is valideval = trueis not valid but
eval = runif(1) > 0.5is valid
Chunk names (or labels)
The chunk below is called plot1.
```{r plot1}ggplot(cars, aes(dist, speed)) + geom_point()```All chunks have a label regardless of whether it is explicitly supplied or not.
Do not include spaces, "_" or punctuation marks in your chunk name!
🔧 Pick your challenge:
challenge-07-include.Rmdchallenge-08-global-option.Rmdchallenge-09-figures.Rmdchallenge-10-cache.Rmdchallenge-11-names.Rmd
25:00
Inline code
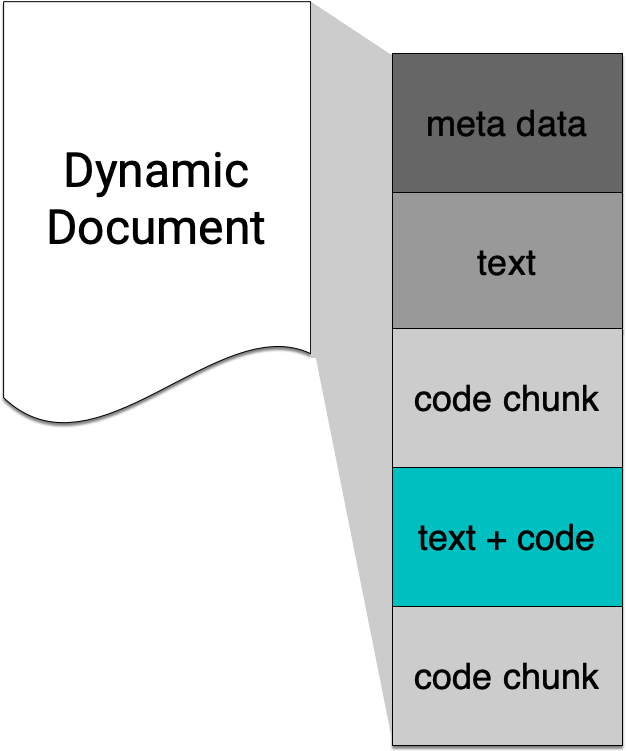
Inline R Commands
Text can contain inline R commands where output is evaluated but code is not echoed.
Text
Updated last: `r Sys.Date()`Output
Updated last: 2019-12-03
Text
The value of $\pi$ is `r pi`.Output
The value of π is 3.1415927.
Accessing YAML meta data inline
All YAML meta data are stored in rmarkdown::metadata as a list.
---title: "R Markdown Workshop"output: html_document---rmarkdown::metadata$title [1] "R Markdown Workshop"All key values of params is stored as a list in params.
---params: data: datav1.csvoutput: pdf_document---params$data [1] "datav1.csv"Stack your Rmd components as you want but only one YAML piece and it needs to be on the top!

For workflow, you'd probably want to keep your tower's height manageable.
R Markdown is not just for R
```{python, echo = FALSE}a = [1, 2, 3]a[0]``` 1```{bash, echo = FALSE}date +%B``` DecemberSharing your html documents
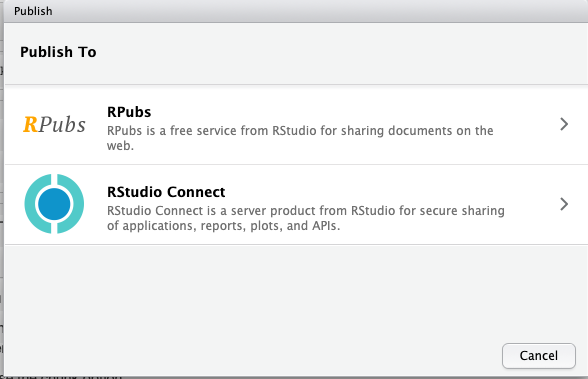
RPubs
After knitting, find this Publish button:

and select RPubs if there is a choice:
Netlify: without Git
- Rename your output html file as
index.html - Go to https://app.netlify.com/
- If you are logged in, go to the bottom of Sites to find
- Drag and drop the folder that contains the
index.html - 🎉 it's alive!
- You probably want to change the site name - to do this go to site > site settings > change site name.
Netlify: with Git
- Rename your output html file as
index.html - Git commit your folder (or html file alone if no dependency)
- Host it on GitHub (my preference), GitLab or Bitbucket
- Go to https://app.netlify.com/ and create a new site from the repo
- 🎉🎉🎉🎉🎉
- Again, you probably want to change the site name - to do this go to site > site settings > change site name.
Happy sharing!
That's it for the basics!
Session Information
devtools::session_info() ─ Session info ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── setting value version R version 3.6.0 (2019-04-26) os macOS Mojave 10.14.6 system x86_64, darwin15.6.0 ui X11 language (EN) collate en_AU.UTF-8 ctype en_AU.UTF-8 tz Australia/Adelaide date 2019-12-03 ─ Packages ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── package * version date lib source anicon 0.1.0 2019-05-28 [1] Github (emitanaka/anicon@377aece) assertthat 0.2.1 2019-03-21 [1] CRAN (R 3.6.0) backports 1.1.4 2019-04-10 [1] CRAN (R 3.6.0) broom 0.5.2 2019-04-07 [1] CRAN (R 3.6.0) callr 3.3.1 2019-07-18 [1] CRAN (R 3.6.0) cellranger 1.1.0 2016-07-27 [1] CRAN (R 3.6.0) cli 1.1.0 2019-03-19 [1] CRAN (R 3.6.0) colorspace 1.4-1 2019-03-18 [1] CRAN (R 3.6.0) countdown 0.2.0 2019-05-30 [1] Github (gadenbuie/countdown@c8e8710) crayon 1.3.4 2017-09-16 [1] CRAN (R 3.6.0) desc 1.2.0 2018-05-01 [1] CRAN (R 3.6.0) devtools 2.0.2 2019-04-08 [1] CRAN (R 3.6.0) digest 0.6.22 2019-10-21 [1] CRAN (R 3.6.0) dplyr * 0.8.3 2019-07-04 [1] CRAN (R 3.6.0) emo 0.0.0.9000 2019-06-03 [1] Github (hadley/emo@02a5206) evaluate 0.14 2019-05-28 [1] CRAN (R 3.6.0) forcats * 0.4.0 2019-02-17 [1] CRAN (R 3.6.0) fs 1.3.1 2019-05-06 [1] CRAN (R 3.6.0) generics 0.0.2 2018-11-29 [1] CRAN (R 3.6.0) ggplot2 * 3.2.1 2019-08-10 [1] CRAN (R 3.6.0) glue 1.3.1.9000 2019-10-24 [1] Github (tidyverse/glue@71eeddf) gtable 0.3.0 2019-03-25 [1] CRAN (R 3.6.0) haven 2.1.0 2019-02-19 [1] CRAN (R 3.6.0) hms 0.5.1 2019-08-23 [1] CRAN (R 3.6.0) htmltools 0.4.0 2019-10-04 [1] CRAN (R 3.6.0) httr 1.4.1 2019-08-05 [1] CRAN (R 3.6.0) icon 0.1.0 2019-05-28 [1] Github (ropenscilabs/icon@a510f88) jsonlite 1.6 2018-12-07 [1] CRAN (R 3.6.0) knitr 1.25 2019-09-18 [1] CRAN (R 3.6.0) lattice 0.20-38 2018-11-04 [1] CRAN (R 3.6.0) lazyeval 0.2.2 2019-03-15 [1] CRAN (R 3.6.0) lifecycle 0.1.0 2019-08-01 [1] CRAN (R 3.6.0) lubridate 1.7.4 2018-04-11 [1] CRAN (R 3.6.0) magrittr 1.5 2014-11-22 [1] CRAN (R 3.6.0) Matrix 1.2-17 2019-03-22 [1] CRAN (R 3.6.0) memoise 1.1.0 2017-04-21 [1] CRAN (R 3.6.0) modelr 0.1.4 2019-02-18 [1] CRAN (R 3.6.0) munsell 0.5.0 2018-06-12 [1] CRAN (R 3.6.0) nlme 3.1-140 2019-05-12 [1] CRAN (R 3.6.0) pillar 1.4.2 2019-06-29 [1] CRAN (R 3.6.0) pkgbuild 1.0.3 2019-03-20 [1] CRAN (R 3.6.0) pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 3.6.0) pkgload 1.0.2 2018-10-29 [1] CRAN (R 3.6.0) prettyunits 1.0.2 2015-07-13 [1] CRAN (R 3.6.0) processx 3.4.1 2019-07-18 [1] CRAN (R 3.6.0) ps 1.3.0 2018-12-21 [1] CRAN (R 3.6.0) purrr * 0.3.2 2019-03-15 [1] CRAN (R 3.6.0) R6 2.4.0 2019-02-14 [1] CRAN (R 3.6.0) Rcpp 1.0.2 2019-07-25 [1] CRAN (R 3.6.0) readr * 1.3.1 2018-12-21 [1] CRAN (R 3.6.0) readxl 1.3.1 2019-03-13 [1] CRAN (R 3.6.0) remotes 2.0.4 2019-04-10 [1] CRAN (R 3.6.0) reticulate 1.12 2019-04-12 [1] CRAN (R 3.6.0) rlang 0.4.0.9000 2019-08-03 [1] Github (r-lib/rlang@b0905db) rmarkdown 1.16 2019-10-01 [1] CRAN (R 3.6.0) rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.6.0) rstudioapi 0.10 2019-03-19 [1] CRAN (R 3.6.0) rvest 0.3.4 2019-05-15 [1] CRAN (R 3.6.0) scales 1.0.0 2018-08-09 [1] CRAN (R 3.6.0) sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 3.6.0) stringi 1.4.3 2019-03-12 [1] CRAN (R 3.6.0) stringr * 1.4.0 2019-02-10 [1] CRAN (R 3.6.0) testthat 2.2.1 2019-07-25 [1] CRAN (R 3.6.0) tibble * 2.1.3 2019-06-06 [1] CRAN (R 3.6.0) tidyr * 1.0.0 2019-09-11 [1] CRAN (R 3.6.0) tidyselect 0.2.5 2018-10-11 [1] CRAN (R 3.6.0) tidyverse * 1.2.1 2017-11-14 [1] CRAN (R 3.6.0) usethis 1.5.0 2019-04-07 [1] CRAN (R 3.6.0) vctrs 0.2.0.9000 2019-08-03 [1] Github (r-lib/vctrs@11c34ae) whisker 0.3-2 2013-04-28 [1] CRAN (R 3.6.0) withr 2.1.2 2018-03-15 [1] CRAN (R 3.6.0) xaringan 0.9 2019-03-06 [1] CRAN (R 3.6.0) xfun 0.10 2019-10-01 [1] CRAN (R 3.6.0) xml2 1.2.0 2018-01-24 [1] CRAN (R 3.6.0) yaml 2.2.0 2018-07-25 [1] CRAN (R 3.6.0) zeallot 0.1.0 2018-01-28 [1] CRAN (R 3.6.0) [1] /Library/Frameworks/R.framework/Versions/3.6/Resources/libraryThese slides are licensed under
 &
& 

